Fully Customizable AI-Powered OAR & Target Segmentation with Automatic Quality Assurance
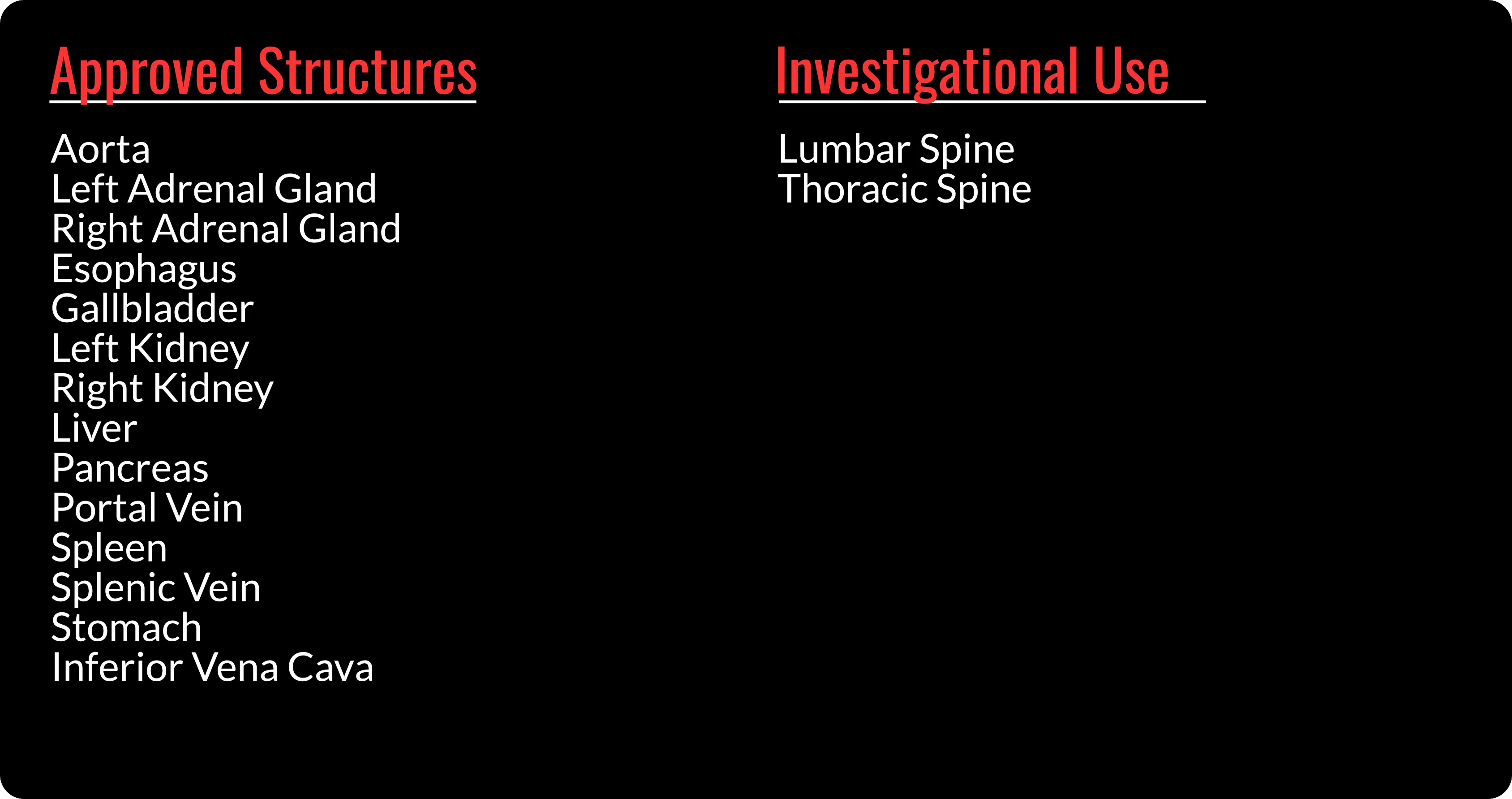
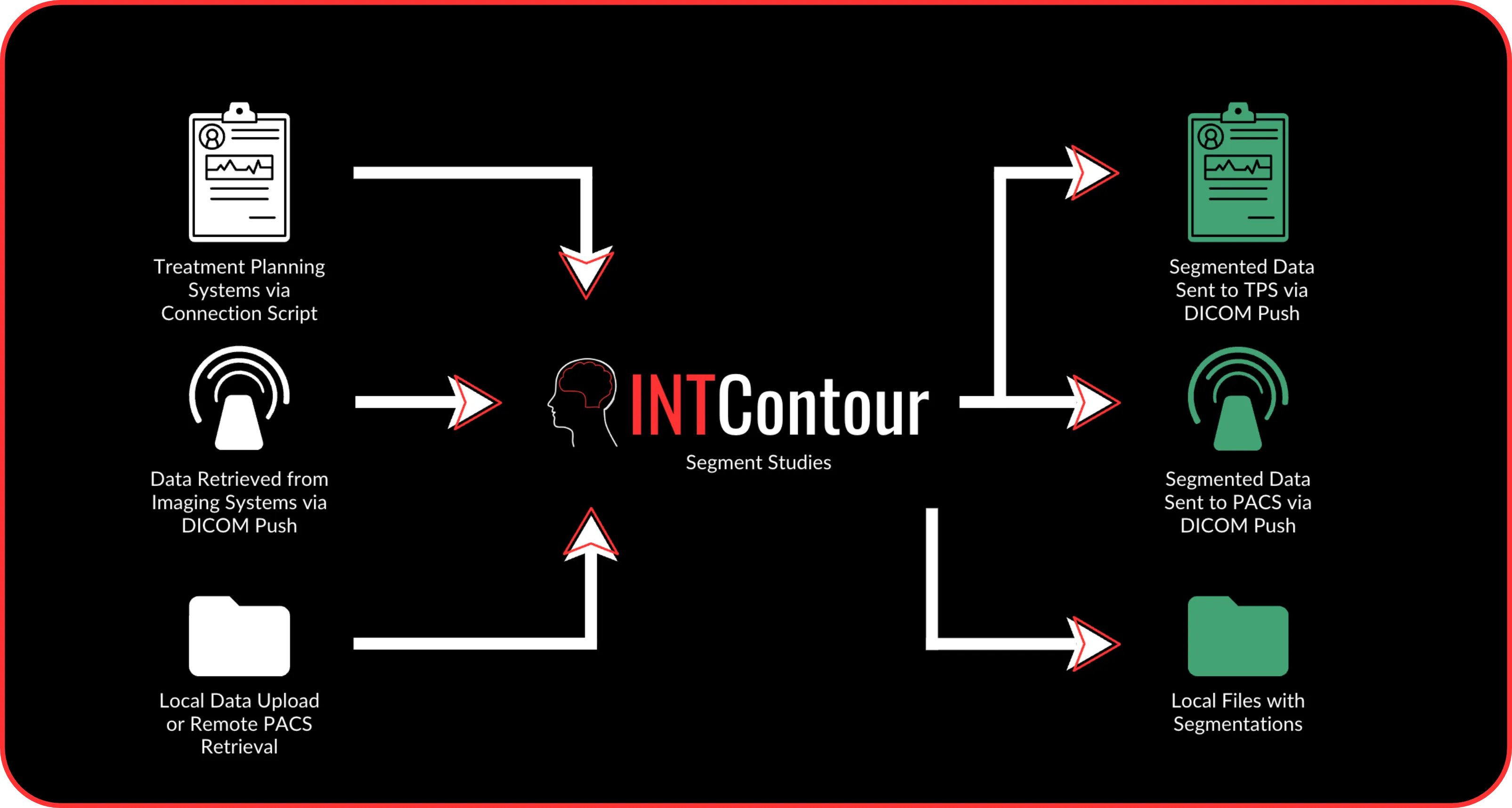
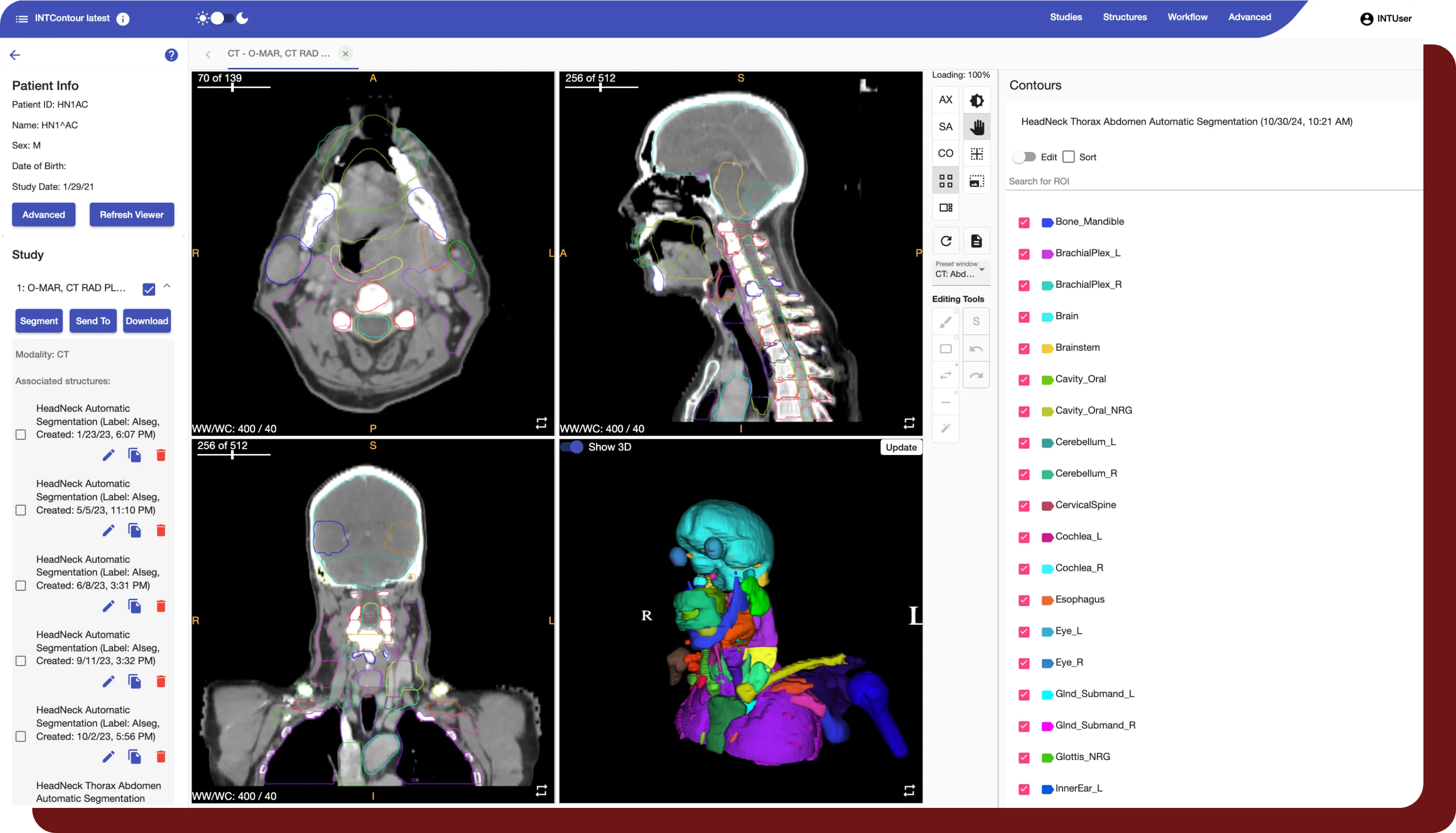
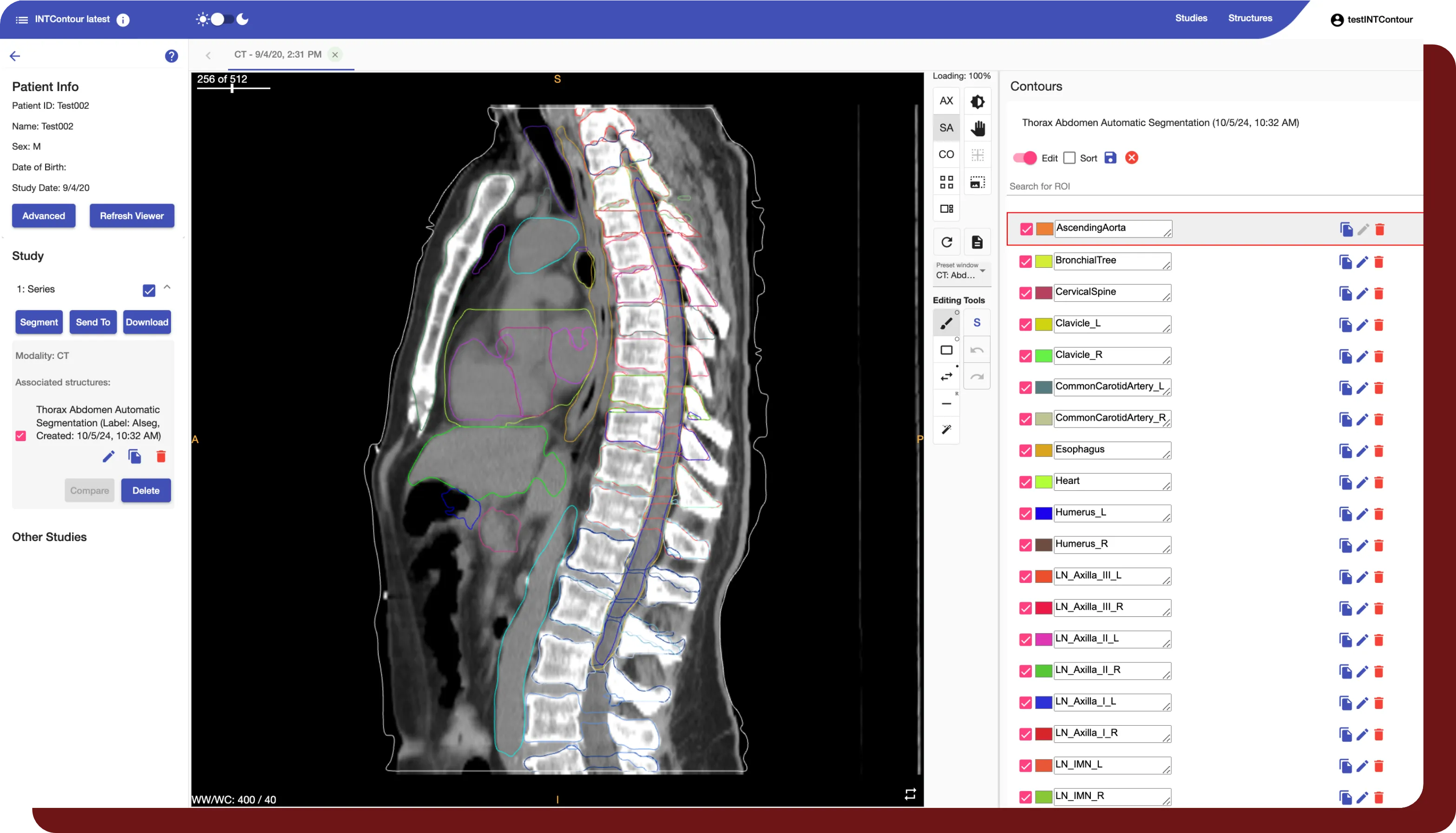
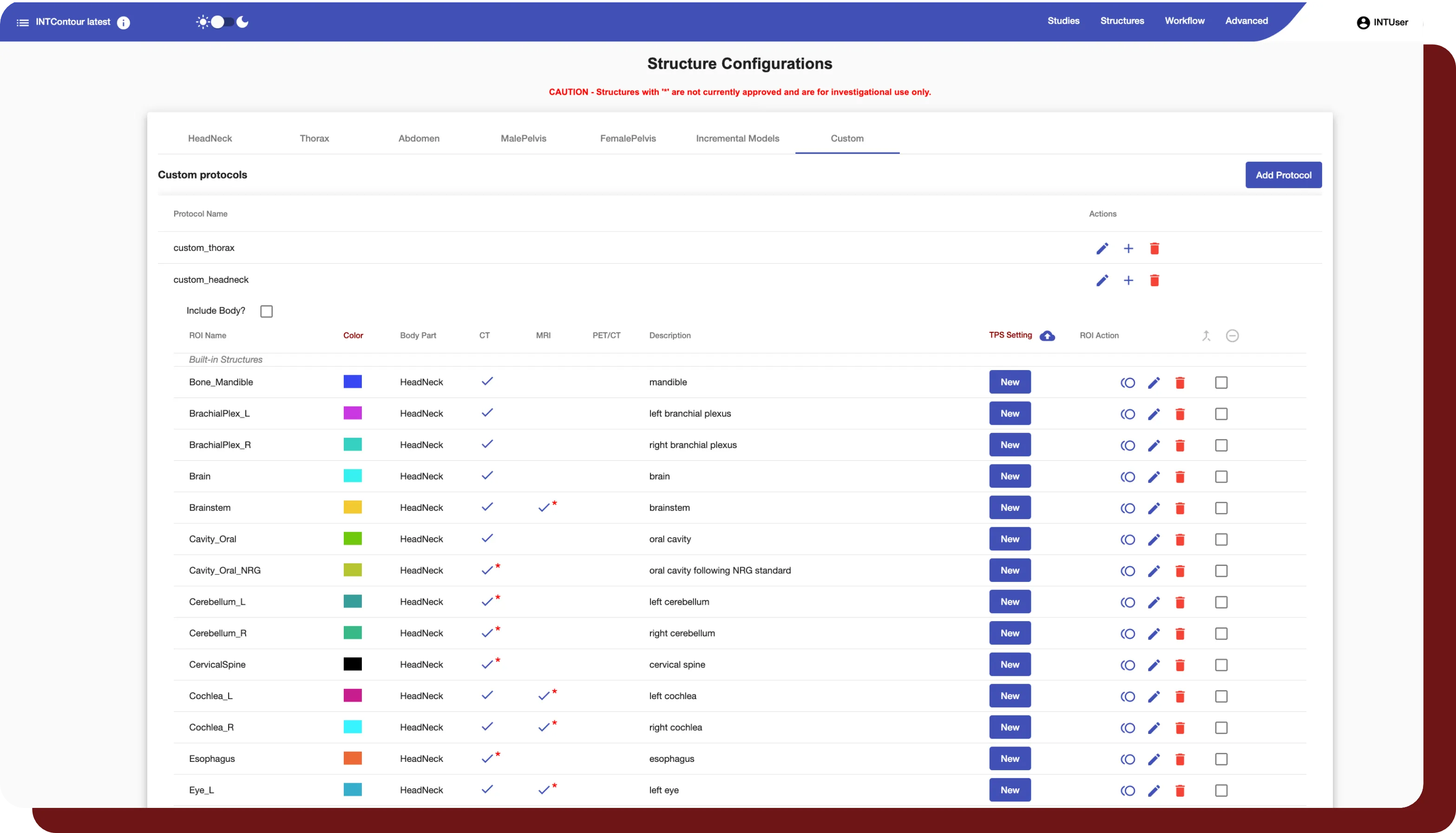
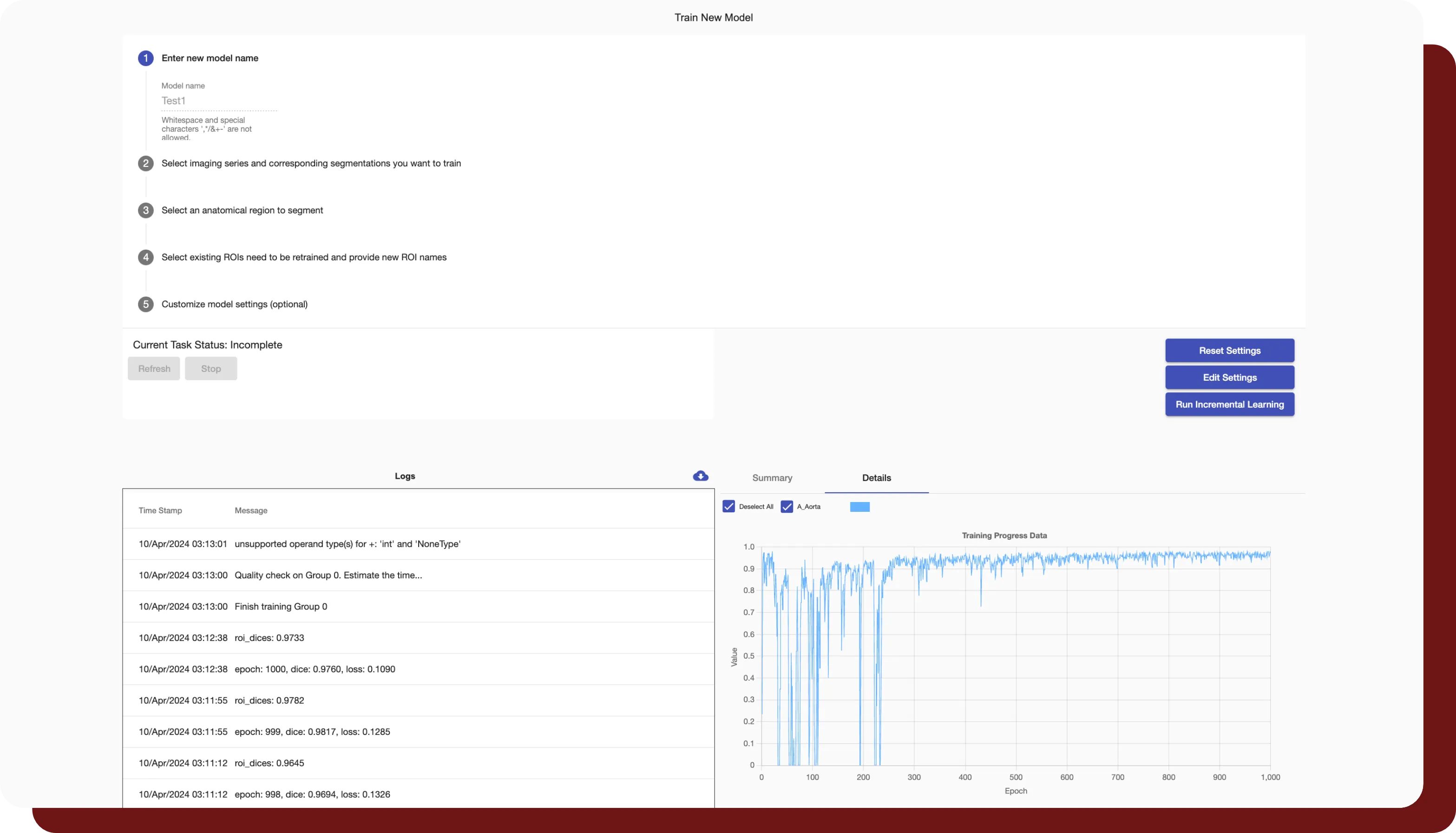
INTContour can automatically segment over 120 organs & lymph node groups from CT, MRI, and PET/CT modalities using built-in or custom protocols. In addition, not only does INTContour provide quality assurance features to allow for segmentation validation, but also provides tools for manual segmenting & annotations, which allow for consistent user control & analysis throughout all studies.